|
Viral Reference Proteomes Help
What are Viral Reference Proteomes?
Viral Reference Proteomes (Viral RPs) are computed from UniProtKB virus complete proteomes. For each pair of proteomes, we calculate their co-membership in UniRef50 clusters. We then hierarchically cluster the similar proteomes into a set of Representative Proteome Groups (RPGs) based on their co-memberships at the cutoff levels of 95%, 75%, 55%, 35% and 15%. The proteomes in each RPG are ranked using a Proteome Priority Score to facilitate the selection of a top ranked proteome as the representative from the group. We also use taxonomic group and host information to annotate the viral proteomes in each RPG. Viral RPs can be used to improve proteome annotation, protein classification, and taxonomic nomenclature bias detection in the viral proteome community.
Viral Reference Proteomes BLAST search
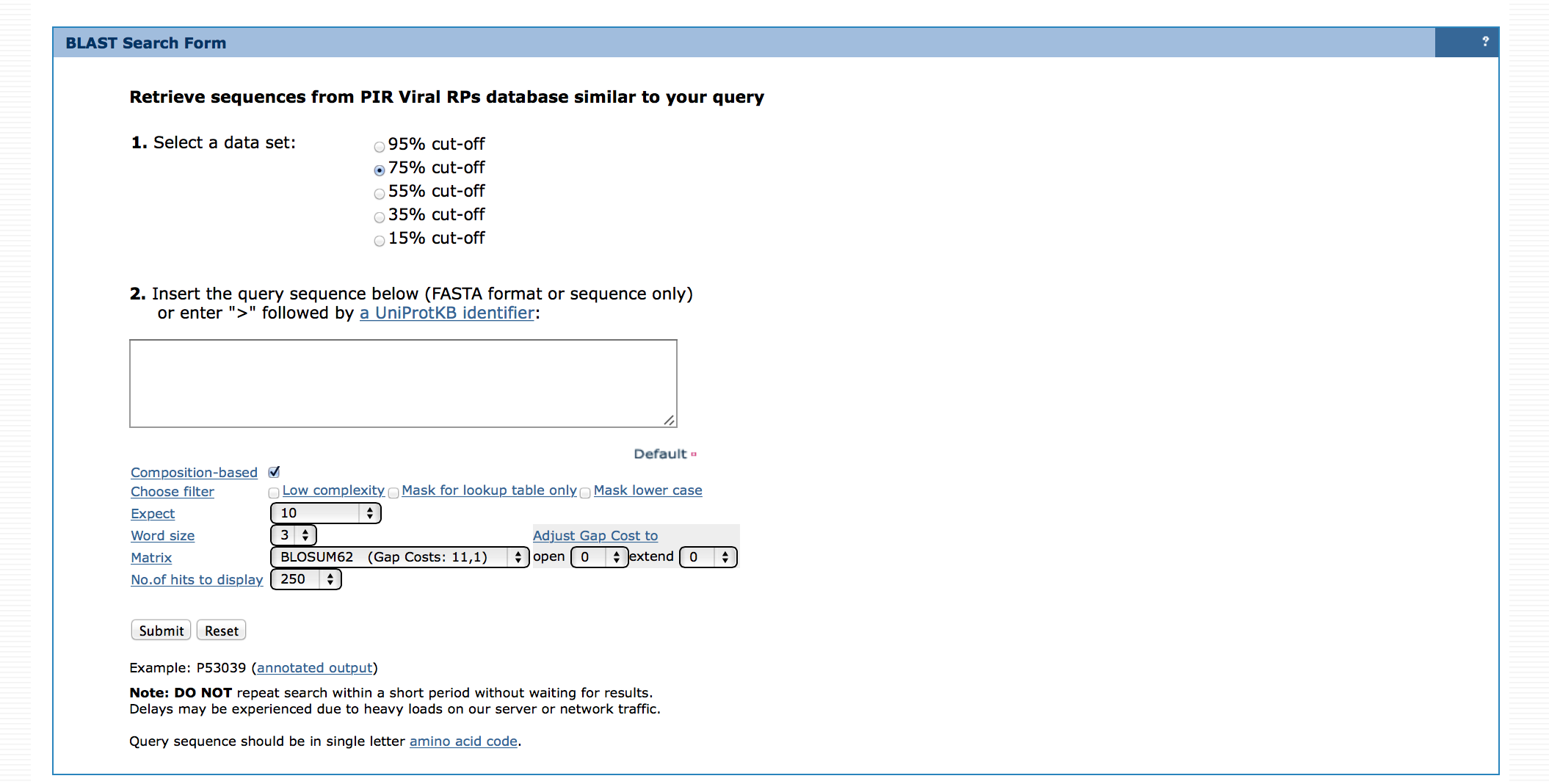
BLAST search can be performed against RP95, RP75, RP55, RP35 or RP15. Lower RPs (such as RP15) has fewer proteomes than higher RPs (such as RP75). For example, to get the least number of BLAST results choose RP15. For additional help on BLAST search please see here.
Browse Viral Reference Proteomes
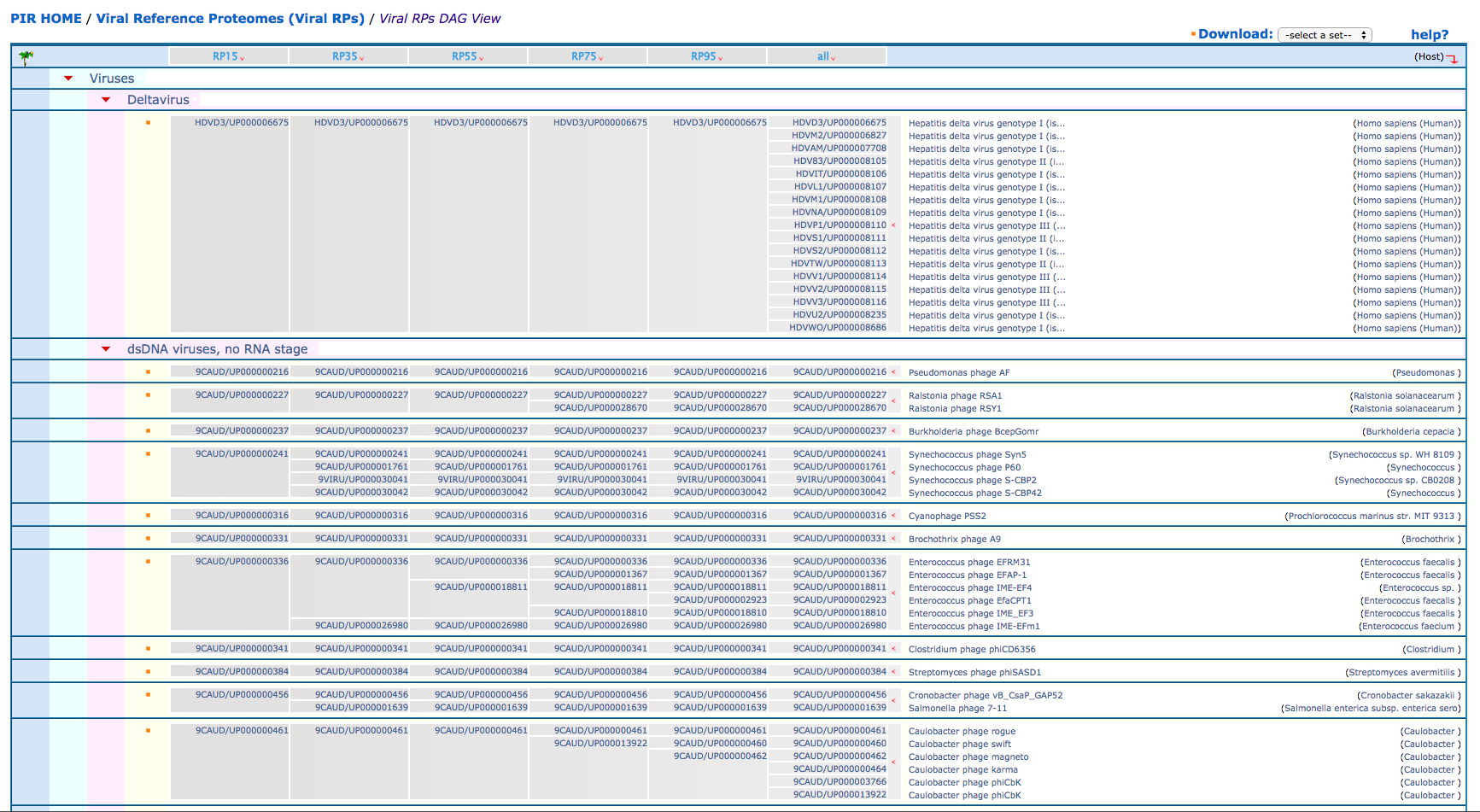
The taxonomic group view of Viral RPs at five different co-membership cutoff levels can be viewed from here. The top most nodes are Deltavirus, dsDNA viruses, dsRNA viruses, environmental samples, Retro-transcribing viruses, Satellites, ssDNA viruses, ssRNA viruses, and Other viruses. The fully expanded view shows all the proteomes that have been analyzed to identify the RPs. Browsing the RPs at different threshold for different taxonomy nodes can provide clues as to which CMT is best for a particular branch and how the RPs are distributed in their taxonomic group tree. Once a desired set of RPs is displayed on the screen, it can be printed for future reference.
Download RPGs and RP Sequences
For the co-membership cutoff levels of 95%, 75%, 55%, 35% and 15%, corresponding Viral Representative Proteome Group files are provided, via links from Viral RP home page in the format below:
>rp_UPId rp_taxon_id rp_oscode rp_name rp_taxon rp_PPS(details) C (CUTOFF) X_to_seed(X-seed)
mp_UPId mp_taxon_id mp_oscode mp_name mp_taxon mp_PPS(details) X_to_rp (X-RP) X_to_seed(X-seed)
Where rp is Reference Proteome, mp is Member Proteome in the RPG.
An example of Representative Proteome Group is shown below:
>UP000000252 374526 9CAUD Lactococcus phage ul36k1t1. dsDNA viruses, no RNA stage 27104.09017(PPS:0,1,1,1.82,53) 75(CUTOFF) 100.00000(X-seed)
UP000000251 374529 9CAUD Lactococcus phage ul36t1k1. dsDNA viruses, no RNA stage 27104.08554(PPS:0,1,1,1.78,51) 86.27451(X-RP) 86.27451(X-seed)
UP000001579 374527 9CAUD Lactococcus phage ul36t1. dsDNA viruses, no RNA stage 27104.08430(PPS:0,1,1,1.76,52) 80.76923(X-RP) 80.76923(X-seed)
We also provide the sequences file in FASTA format for the RP95, RP75, RP55, RP35, and RP15 sets. In addition, users can choose to make their own customized RP set by using the taxonomic group based table or perl sscript available via a link from the Viral RP home page.
Customizable RP Sequences
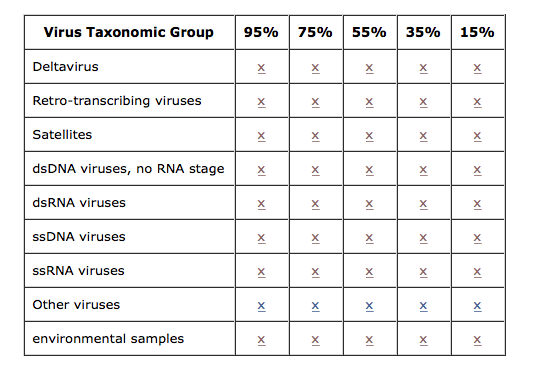
We provide two ways to allow users create customized RP sequences file with respect to Virus taxonomic groups and co-membership cutoff levels.
- User can run the following PERL script with a configuration file customized to his/her needs to download the RP sequences.
Perl Script
#
# Make your own RP sequence file
#
# This script will create a RP sequence file using the user's choices of
# Taxonomy Groups and RPG cutoffs.
#
# The perl module "LWP::Simple" is required
#
#!/usr/bin/perl
use LWP::Simple;
if(@ARGV != 2) {
print "Usage: perl getMyRPSeq.pl mypick.txt output.seq\n";
exit 1;
}
# configuration file of tab-delimited txt file with three columns:
# TaxonGroup TaxonId RPCutoff
# Note: for each row in the file, only the third column should be changed.
# The possible values are 15, 35, 55, 75, 95
my $mypick = $ARGV[0];
# path to the output sequence file
my $output = $ARGV[1];
my %taxonId = ();
my $url = 'http://pir.georgetown.edu/rps/viruses/data/rp_seq_tax_group/current/';
open(PICK, $mypick) or die "Can't open $!\n";
while($line=) {
chomp($line);
if(!($line =~ /^Virus Taxonomic Group/ || $line =~ /^#/ || $line =~ /^$/)) {
my ($taxId, $cutoff) = (split(/\t/, $line))[1, 2];
my $seqUrl = $url."/".$cutoff."/".$taxId.".seq";
my $file = $taxId.".seq";
$taxonId{$taxId} = 1;
my $status = getstore($seqUrl, $file);
die "Error $status on $seqUrl" unless is_success($status);
}
}
close(PICK);
open(OUT, ">", $output) or die "Can't open $!\n";
for my $key (sort keys %taxonId) {
my $seqFile = $key.".seq";
if(-e $key.".seq") {
open(FH, $seqFile) or die "Can't open $!\n";
while($line=) {
print OUT $line;
}
close(FH);
unlink($seqFile);
}
}
close(OUT);
Configuration File
# configuration file of tab-delimited txt file with three columns:
# VirusGroup TaxonId RPCutoff
# Note: for each row in the file, only the third column should be changed.
# The possible values are 15, 35, 55, 75 95
#
Virus Taxonomic Group Id RPCutoff
Deltavirus 39759 75
Retro-transcribing viruses 35268 75
Satellites 12877 75
dsDNA viruses, no RNA stage 35237 75
dsRNA viruses 35325 75
ssDNA viruses 29258 75
ssRNA viruses 439488 75
Other viruses 10239 75
environmental samples 186616 75
Download the RP sequences using Virus taxonomic group and co-membership cutoff levels information by getting one file from each row and then put them together to form the customized RP sequences file.
|


